In any Knowledge Graph-based project, keeping track of where data comes from is important. When you know the source of your facts or assertions, you can contextualize those facts: how relevant is assertion X to my current research, is it from a source that I personally trust, and if I have two conflicting views how can I decide which source to go with? Apart from issues of trust and confidence, tracking the source also can serve more mundane goals, such as knowledge graph maintenance: source X has published a new edition of their data set, so we need to replace the relevant data in our own Knowledge Graph, and so on.
Keeping track of the source of data is often referred to as provenance. In this blog post, we will look at provenance tracking in RDF Knowledge Graphs using the Wikidata dataset as an example, and we will look at how RDF-star and SPARQL-star, two new community efforts to extend the RDF model, can make this task easier.
Source tracking for statements in Wikidata
Wikidata is a massive free and open knowledge base, using the RDF data model to represent knowledge as a graph. It is the structured data equivalent of Wikipedia and other topical wiki systems (see also the metaphactory wikidata demo system).
The RDF data model is a natural choice for capturing this kind of massive heterogeneous data: it is flexible and extensible, open to change and handling exceptions to the rule, and due to its minimal assumptions on schema conformance puts little or no constraints on parallelization (and therefore, scalability) of storage, transformation and extraction solutions.
Let's look at a particular statement in Wikidata: COVID-19 has an effect called "cytokine release syndrome". This is captured in Wikidata by the following RDF statement:
wd:Q84263196 wdt:P1542 wd:Q3961647 .
This is somewhat cryptic perhaps (if you wish to know more about how the Wikidata data model works see their Primer), but Wikidata contains human-readable labels for each of these codes. If we apply those labels, we get the following:
"COVID-19" "has effect" "cytokine release syndrome" .
Or depicted as a graph:
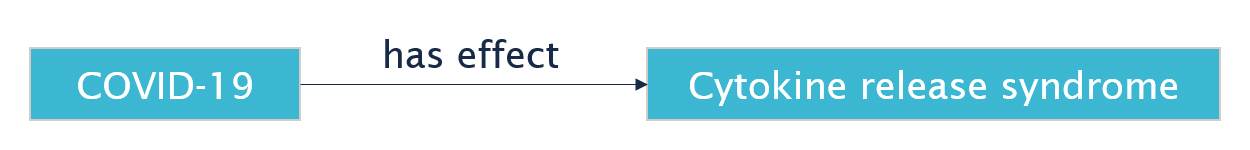
Now suppose that we want to express that this statement about an effect of COVID-19 was asserted in a particular medical publication, namely the article "Effective Treatment of Severe COVID-19 Patients with Tocilizumab" (wd:Q87755922). How can we capture that in RDF?
Presumably, we want our graph to look something like this:

That is, we want to add a property (e.g. "stated in" - wikidata code pr:P248) that starts from an edge in our graph, rather than a node. In other words, we need what is referred to as an edge property in graph databases. Unfortunately, regular RDF has no direct support for this kind of edge property, and this is exactly the kind of thing that RDF-star aims to make it easier to deal with.
Annotating edges in standard RDF
Before RDF-star came along, the lack of edge property annotation capability was already recognized as a problem, and several modeling solutions to deal with it have been devised over the years. Without claiming to give a complete overview, we will briefly discuss a few of these approaches and their pros and cons.
RDF reification
Reification is a general term that roughly means "to turn something abstract into something concrete". In the case of RDF, reification of statements is used to turn the abstract concept (the statement) into something that can be concretely addressed. Put more simply: reification is about making it possible to assert statements about statement.
The RDF standard has reification features built-in. At first glance, this looks as if it is made to order for our problem of stating the source of our statement about COVID-19: after all if we can turn the statement “COVID-19 has effect cytokine release syndrome” into a reified object that we can then say something about (such as "this is stated in article so-and-so"), we’ve achieved our result, right?
Well, yes and no. Standard RDF reification works by creating an additional set of RDF triples that encode the reified object. Our original triple was this:
wd:Q84263196 wdt:P1542 wd:Q3961647 .
Or to use human-readable pseudo identifiers (which I will use from now on):
:COVID19 :has_effect :cytokine_release_syndrome .
In reified form, that becomes this:
:statement1 a rdf:Statement;
rdf:subject :COVID19;
rdf:predicate :has_effect;
rdf:object :cytokine_release_syndrome .
And we can use that to assert our additional provenance fact:
:statement1 :stated_in :effective_treatment_article .
Seen as a graph, we get this:

While this works and has the nice property of being fully compliant with existing RDF standards (so no need for any custom extensions), it has the major downside that it is very verbose: we are introducing 4-5 additional RDF triples for each statement that we want to say something about. Imagine having to do this for every statement in even a medium-sized dataset: you are effectively quadrupling your dataset size.
In addition, RDF reification is difficult to maintain and work with in SPARQL: what happens with the reified statement if the original statement is removed? What happens if part of the statements making up the reification is missing?
For these reasons, RDF reification has seen low uptake as a solution for edge properties.
Named graphs
One other existing approach we will discuss here briefly is named graphs. Named graphs are a feature introduced as part of the SPARQL standard. Put simply it is a mechanism to assign an identifier (a "graph name") to a collection of RDF triples, and then use that identifier as part of SPARQL querying to select certain subsets of RDF data within a larger dataset. It is widely used as a mechanism to keep track of the sources that statements came from, and to organize and partition sets of RDF data that logically "belong together".
While named graphs are about assigning identifiers to collections of statements, a collection of 1 statement is of course possible, which means we can use the named graph mechanism to assign an identifier to each statement where we wish to annotate the edge. For example, if we add our fact about COVID-19 to a named graph:
:namedGraph1 { :COVID19 :has_effect :cytokine_release_syndrome . }
We can then add our "stated in" provenance property as follows:
:namedGraph1 :stated_in :effective_treatment_article .
In graph form, this looks like this:
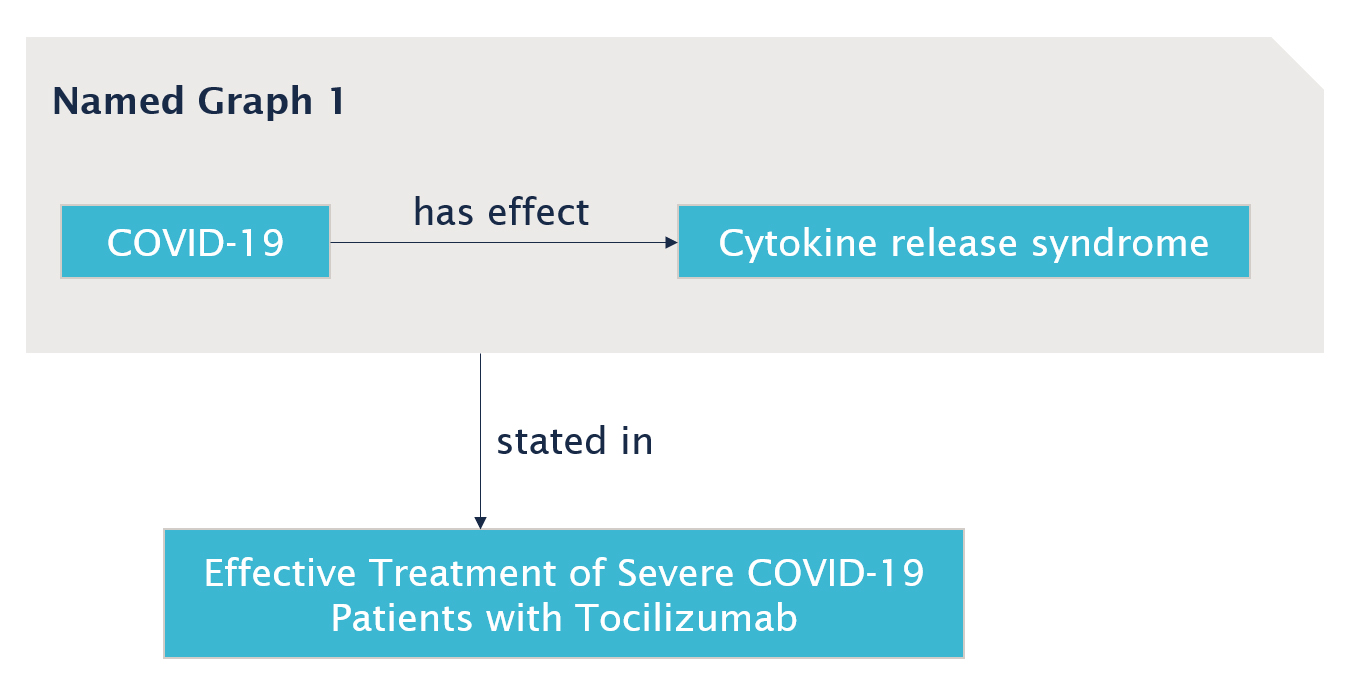
This is an attractive solution because it gives us the flexibility to annotate any relation we wish without being forced to change the core model. Hower, there are some drawbacks to this approach as well. First of all, since named graphs are intended for collections of triples, there is no built-in mechanism to stop us from adding two or more statements to the same named graph, leading to inconsistencies and other possible maintenance issues. An additional practical problem is that most RDF databases are designed with a "reasonable" assumed limit on the total number of named graphs in any dataset, that may be reached sooner when using the mechanism to create a new named graph for every triple in the dataset.
The RDF-star approach
RDF-star deals with the conundrum in a different way: it introduces an extension of the standard RDF conceptual model, as well as some extensions to concrete RDF syntax formats, like Turtle. As an example, the statement about COVID-19 can be embedded in Turtle-star syntax as follows [1]:
<< :COVID19 :has_effect :cytokine_release_syndrome >>
:stated_in :effective_treatment_article .
The double angle brackets that surround the original statement are a new RDF-star feature. They effectively turn the entire statement into a resource (called an embedded triple), that then can be used in turn to make a statement about (in this case, that it was stated in a particular article).
It also introduces an alternative syntax, using a more annotation-like form:
:COVID19 :has_effect :cytokine_release_syndrome
{| :stated_in :effective_treatment_article |} .
Here, the {| … |} markers encapsulate the edge property annotation that applies to the immediately preceding triple.
In addition, RDF-star encompasses an extension of the SPARQL query language, called SPARQL-star. Any RDF database that is RDF-star-enabled can process and interpret data in Turtle-star syntax, and can answer SPARQL-star queries.
As an example, to ask "which effects of COVID-19 were stated in the article "Effective Treatment of…":
SELECT ?effect
WHERE { << :COVID19 :has_effect ?effect >> :stated_in :effective_treatment_article . }
A major advantage of the RDF-star approach is that it is fit for purpose. It takes away the modeling complexity burden for ontology developers by providing a simple mechanism for tagging/annotating individual statements. It also gives RDF database implementors the means to process such annotations in an efficient manner. Many triplestore implementations already are starting to support dedicated indexes to RDF-star triples, which make the approach much more scalable and performant than classic reification. In addition, the syntax is pleasantly compact and easy to work with as a data author, and intuitive to query. It also does not suffer from the drawback of incremental addition of annotations that we have seen in some other approaches: there is a natural way of adding RDF-star annotations to existing data without needing to convert that data into a different form first.
RDF-star in practice: metaphactory, RDF4J and GraphDB
In metaphactory, we are leveraging support for RDF-star that was developed in partnership with the Ontotext GraphDB team and contributed to the open-source RDF4J platform, as well as some major contributions to the SPARQL.js JavaScript library to make processing of RDF-star possible in frontend platforms. The support added to these projects consists of parsers and writers for extended RDF syntax formats such as Turtle-star, as well as (in the case of RDF4J) extensions to the query algebra and underlying query engine, and adding touch points for triplestore vendors to "hook in" in a gradual, making triplestores able to process RDF-star even when not natively supported. This is achieved by doing an internal conversion to use classic RDF reification, so it comes at a cost of scale and performance, but it’s a stepping stone to full native integration (where the triplestore itself can accept embedded RDF-star triples as values in their own right and store and index them efficiently).
The support for RDF-star available in RDF4J enables us to add immediate support for retrieving and displaying edge property data in metaphactory. For example, to display the stated effects of COVID-19 and the sources of those statements, you could use a metaphactory table component, like so:
<semantic-table
query='
PREFIX wd: <http://www.wikidata.org/entity/>
PREFIX wdt: <http://www.wikidata.org/prop/direct/>
PREFIX pr: <http://www.wikidata.org/prop/reference/>
SELECT DISTINCT ?effect ?source
WHERE {
<< wd:Q84263196 wdt:P1542 ?effect >> pr:P248 ?source .
}
'
options='{
"showFilter":false
}'>
</semantic-table>
Which would be rendered as follows:

On a more advanced level, metaphactory can visualize RDF-star annotations graphically. For example, the following diagram displays the provenance information directly as an additional label on the edge between the COVID-19 node and the effect node:
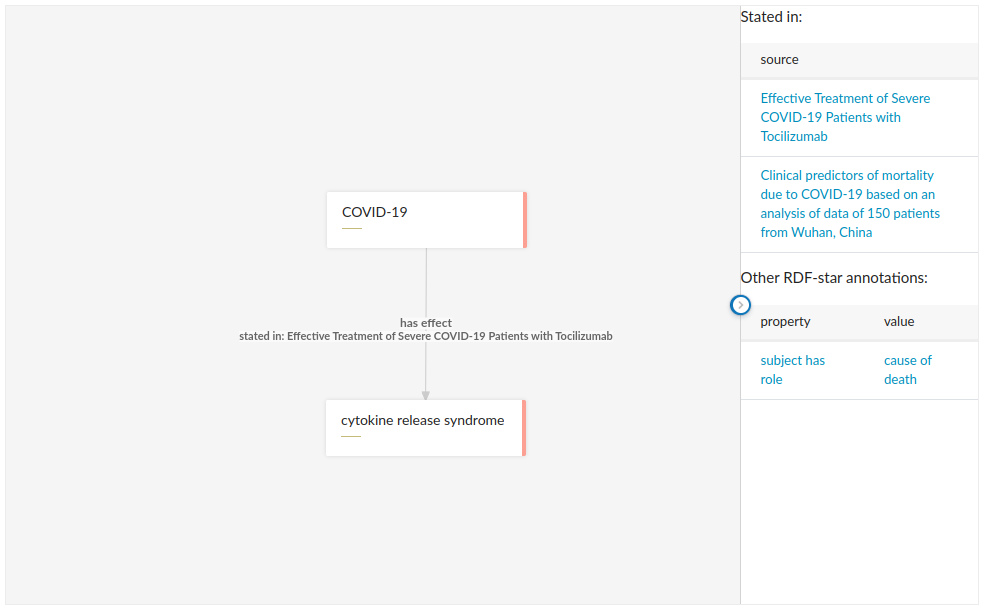
RDF-star in metaphactory
In addition, by clicking on the edge, the diagram will show any RDF-star annotations in the knowledge panel on the right. Note that we have full control over what gets displayed where: in this case, we chose to show only one source annotation on the edge itself, for brevity. However, in the knowledge panel we show full details of all annotations on the selected edge, focusing on source annotations but also leaving room for any other annotations.
These are relatively basic use cases, but what we can already see is that with a very simple extension we can dramatically extend the expressive capabilities of the ontology: we can very easily inject, query, visualize and manipulate provenance information as well as other types of edge annotations.
You can see all this in action on our live demo system at https://rdfstar.metaphacts.cloud/, or if you prefer, have a look at our online help at https://help.metaphacts.com/resource/Help:RDF-star and configure your own metaphactory system to work with RDF-star annotations.
The nice thing about standards
RDF-star is not yet a real standard. At the time of writing, it is a draft proposal from a community group consisting of various stakeholders, including academia and industry. This group was born out of the realization that several existing implementations of RDF-star existed, yet all had differences in the details of that implementation, leading to no two systems being able to interoperate at the level of RDF-star (that is, use the same syntax formats, the same queries, or even the same semantic interpretation).
The goal of the group is to come up with a consensus proposal that implementers can work with, and to reach a level of interoperability between the various implementations. If all goes well and uptake is sufficient, the proposal will hopefully be used as input to a future W3C working group to standardize it.
At metaphacts we are actively taking part in this community group, in particular trying to reach consensus on syntax formats for interoperability, so that systems such as metaphactory, which routinely interact with a variety of triplestore implementations, can confidently manipulate RDF-star data and use SPARQL-star queries without having to have extensive customization for each particular store.
Get in touch to find out more
Get in touch with us to learn more about the possibilities of RDF-star for annotation and make sure to also subscribe to our newsletter or use the RSS feed to stay tuned and learn about future developments on this matter. To get started with metaphactory, sign up for a free trial and don't hesitate to get in touch with us if you have any questions.
[1] I am simplifying how provenance information is captured in Wikidata in reality. The point I am trying to make is not about the specific modeling approach that Wikidata itself has chosen, only about how we can use RDF-star to achieve a simple form of edge property annotation for provenance purposes.
The work reported here is partiallly funded through the European research project FROCKG.



